ACDLabs
ACDLabs

 ACDLabs
ACDLabs home > DRUG DISCOVERY > ACDLabs
home > DRUG DISCOVERY > ACDLabs
스펙트럼, 크로마토그래피 데이터 처리/분석/관리, ADMETox 예측
ACD/Lab사의 가장 기본이 되는 프로그램으로서 하나의 플랫폼으로 NMR, GC/LC/MS, UVIR, Chromatography 등 다양한 분석데이터들을 처리/분석 할 수 있으며, 다양한 분석 방법을 제공하는 글로벌 주요 장비 회사들의 각종 데이터 형식을 지원합니다.

사용이 편리한 GUI를 제공하여 프로그램을 직관적으로 사용할 수 있습니다.
화합물 구조 정보 입력 후 스펙트럼 분석 정보를 이용하여 자동으로 peak assign을 해주고, 일상적인 chemical structure confirmation
작업을 손쉽게 수행할 수 있습니다.
Report template 제작 툴을 이용하여 원하는 양식의 report format을 만들 수 있고, report 생성 버튼 클릭으로 필요한 report를 생성할 수 있습니다.
NMR Predictors Suite
The most Accurate 1D and 2D NMR Predictors
화합물 구조로부터 빠르고 정확하게 chemical shifts, coupling constants 등 1H, 13C, 15N, 19F, 31P 핵종에 대한 NMR spectra를 예측할 수 있습니다.
In house data들을 trainable database에 추가하여 예측 신뢰도를 높일 수 있습니다.
효과적으로 structure verification할 수 있도록 예측 결과와 실험 결과를 직접 비교해 볼 수 있습니다.
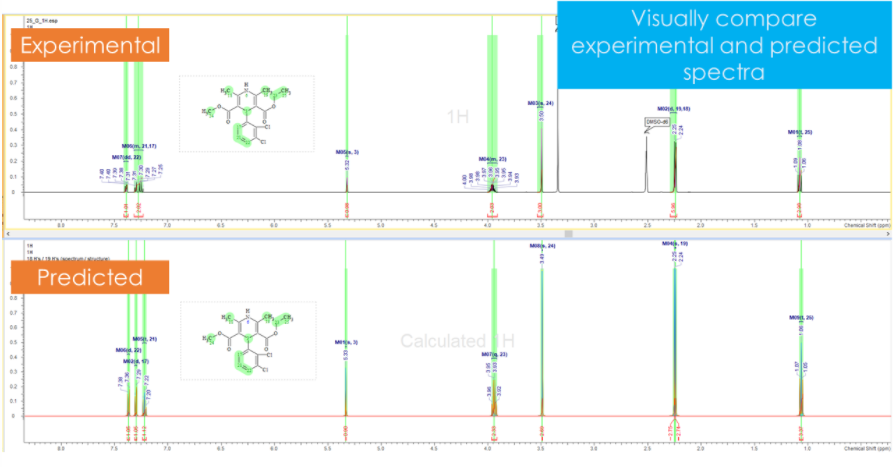
- 다양한 용매 조건 하에서 1H, 13C NMR spectra 예측
- 예측 시 tautomeric forms 인식 및 제안
- 15N chemical shifts를 예측하고 acquisition range를 조절하여 측정 시간을 단축할 수 있습니다.
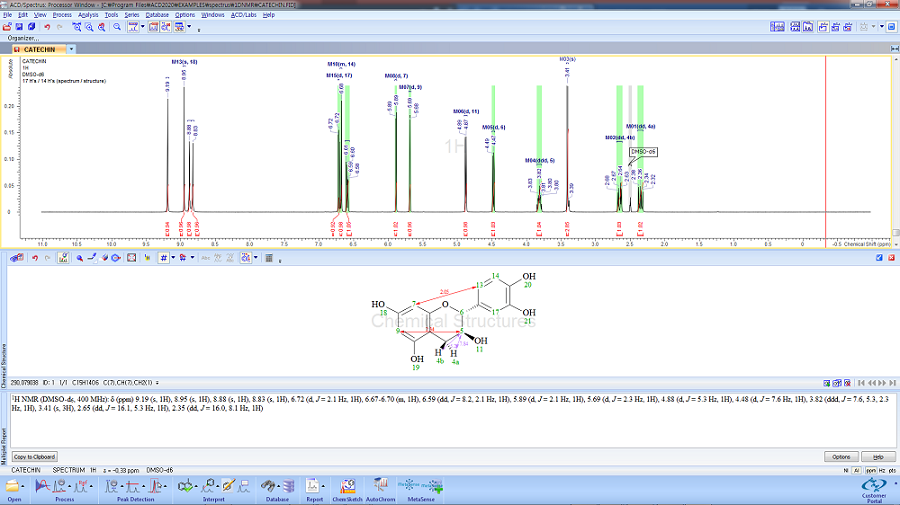
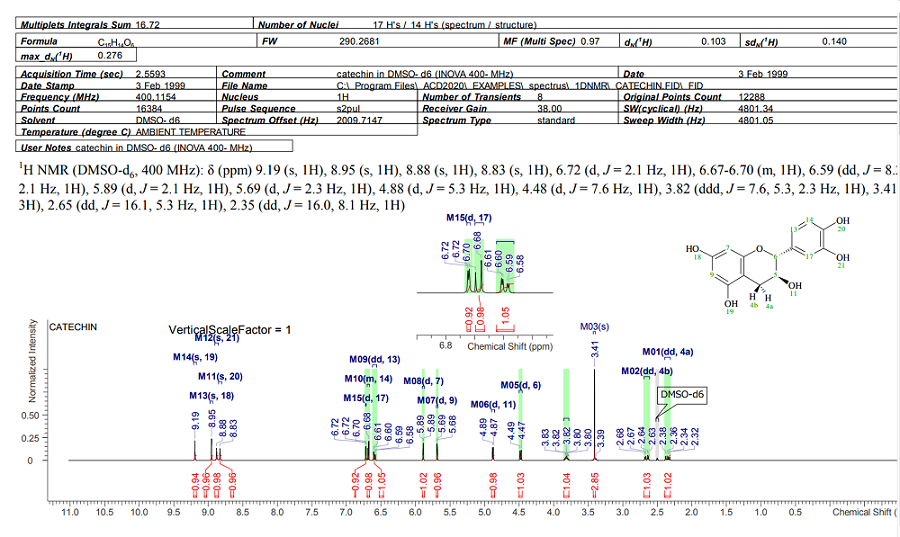
NMR Workbook Suite
Data Interpretation, Structure Characterization & Knowledge Management
NMR Project 기능을 통해 1D, 2D NMR을 동기화 하여 피크 선택 시, 1D, 2D 모두에 반영됩니다.
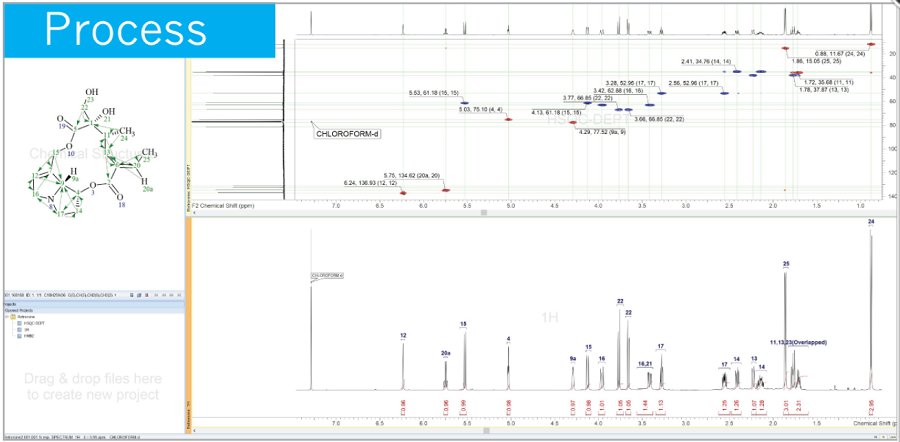
모든 형식의 기본 또는 사용자 정의 보고서 템플릿을 사용하여 전문적이고 고품질의 보고서를 생성할 수 있습니다.
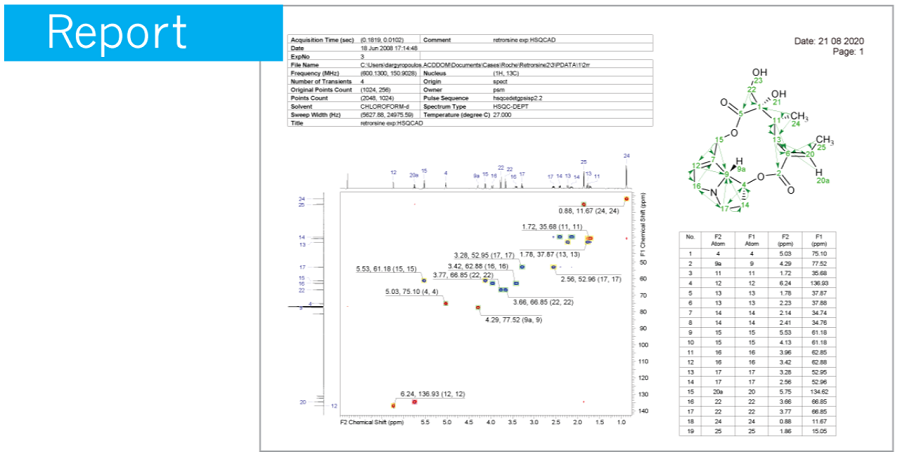
한 번의 간단한 단계를 거쳐 실험 샘플과 관련된 모든 실험 스펙트럼들을 데이터베이스로 구축 할 수 있습니다.
개별 데이터베이스 기록에는 화학구조 및 다양한 분석 결과가 포함될 수 있습니다.
(table of peaks, table of assignments, annotations, etc.)
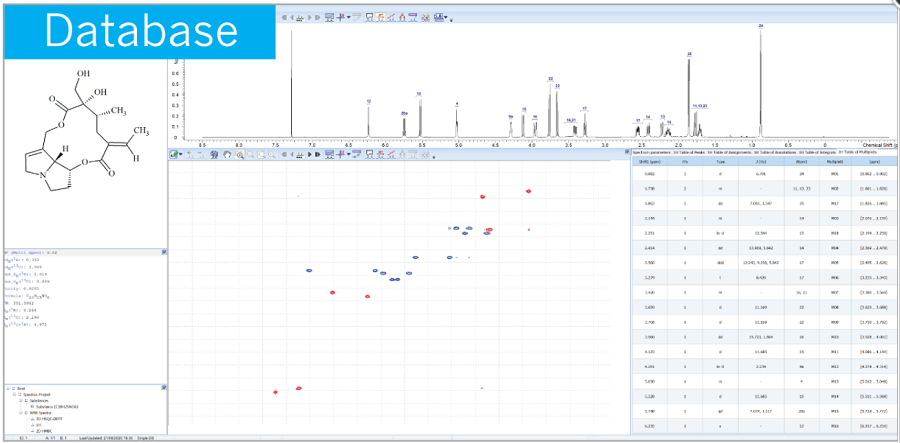
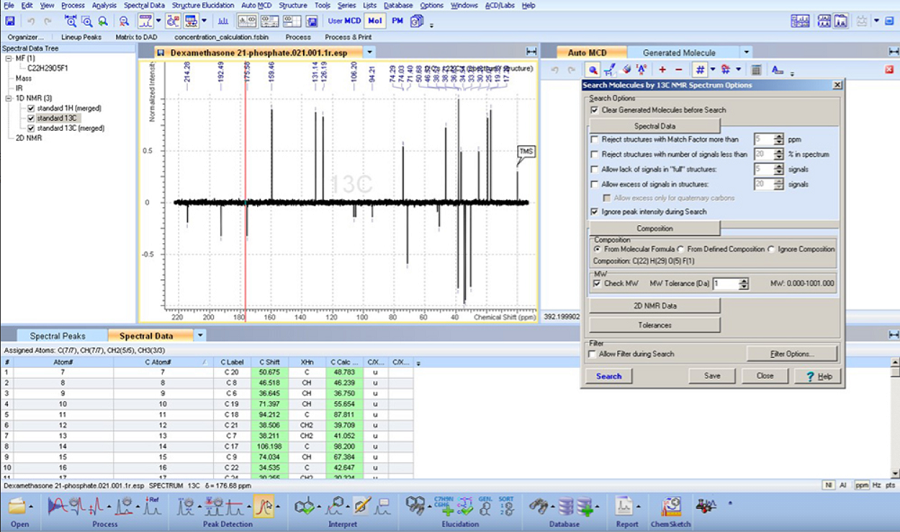
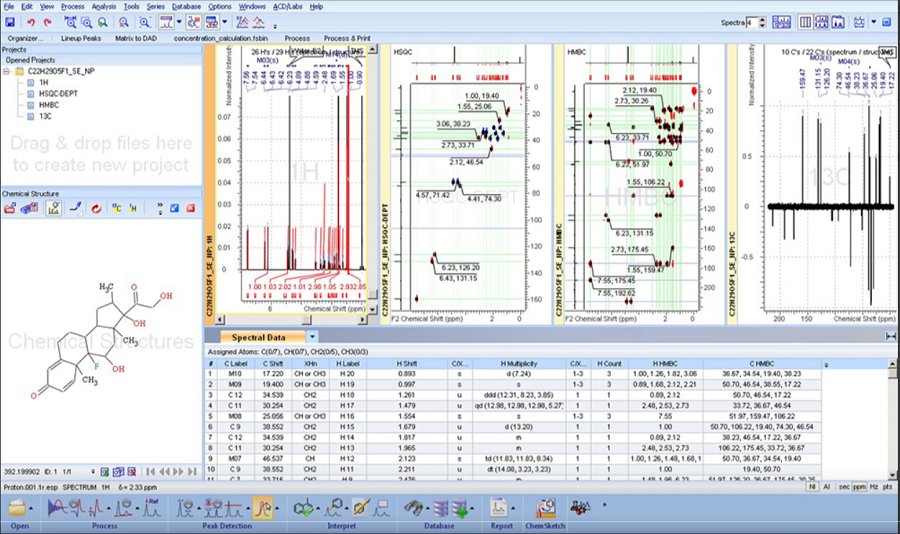
Structure Elucidator Suite
Rapid Dereplication and de novo Elucidation for Complex Unknown Structures
기존에 알려진 1H, 13C NMR database와 약 1억개에 달하는 예측된 1H, 13C NMR database를 빠르게 검색하여 known/unknown structure를 분별할 수 있습니다.
NMR과 다른 분석 기술들을 사용하여 수준 높은 구조 검증과 신속한 구조 특성화가 가능하고, 복잡하고 구조가 알려지지 않은 물질들의
구조를 해명할 수 있습니다.
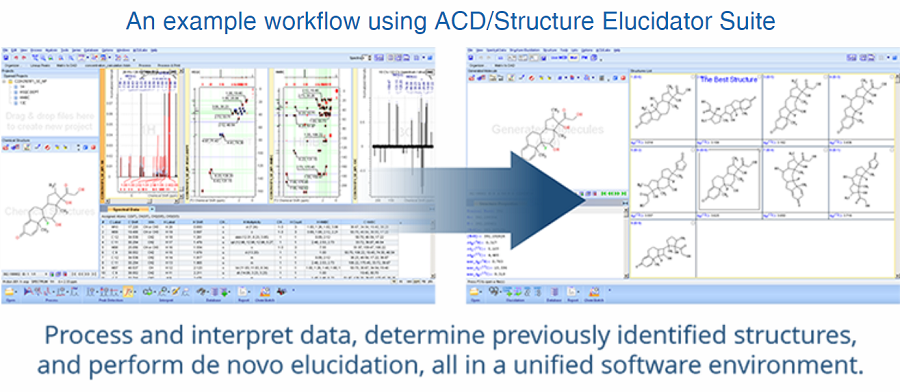
Dereplication: 기 구축된 NMR 라이브러리의 스펙트럼 검색으로, 스펙트럼과 관계된 이미 밝혀진 구조 정보를 빠르게 검색할 수 있습니다.
Elucidation: 관측된 1D, 2D NMR spectra를 만족하는 구조들을 생성합니다.
Name
Chemical Names according to IUPAC and CAS Index rules
IUPAC 및 CAS 인덱스 규칙에 따라 화학적 이름을 생성하고 이름을 구조로 다시 변환할 수 있으며, 생물학적 분자, 유기 금속 및 폴리머와
같은 까다로운 명명 영역을 쉽게 처리 할 수 있습니다.
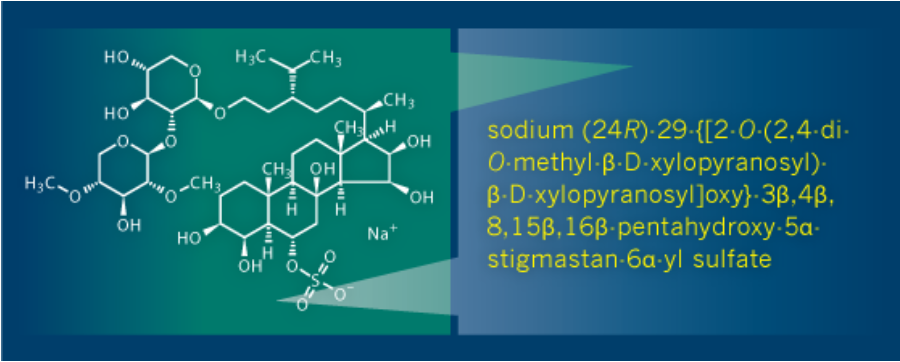
Generate the IUPAC name for your structure, or enter a name to produce the chemical structure
MS Fragmenter
Estimating Mass Spectral Fragmentation
구조를 그리고, 이온화 방법 및 극성을 선택하여 실시간으로 MS fragmentation을 예측할 수 있습니다.
표적 분자 조각화에 대한 통찰력을 얻기 위해 확립된 문헌 규칙에 따라 조각 이온을 예측합니다.
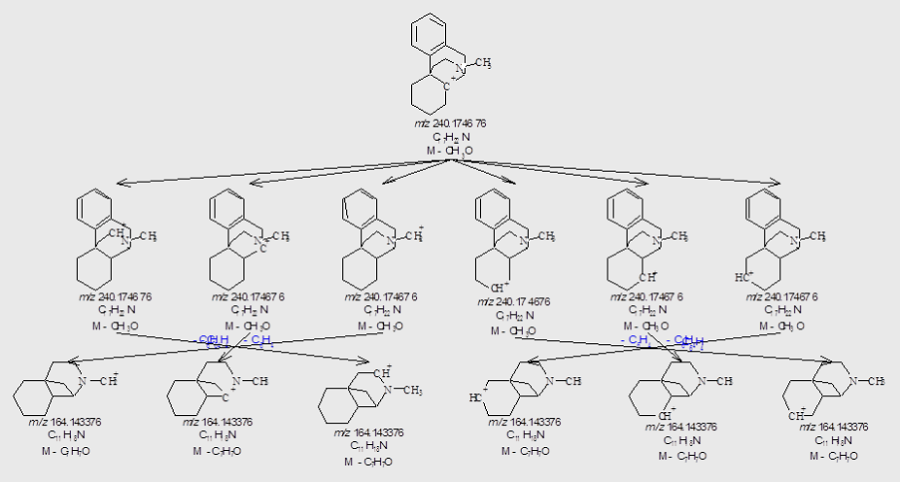
MS Workbook Suite
Detecting, Identifying, and Characterizing LC/MS & GC/MS Components
LC/UV/MS & GC/MS 데이터를 사용하여 화합물을 식별하고 특성화하기 위한 전문적인 툴입니다.
Agilent, Bruker, LECO, PerkinElmer, SCIEX, Shimadzu, Thermo Fisher Scientific, Waters 등 대부분의 주요 장비 공급업체의 파일 포맷을
인식합니다.
스펙트럼, 크로마토 그래피 및 구조 라이브러리와 맞춤형 보고서를 생성하여 지식을 효율적으로 공유합니다.
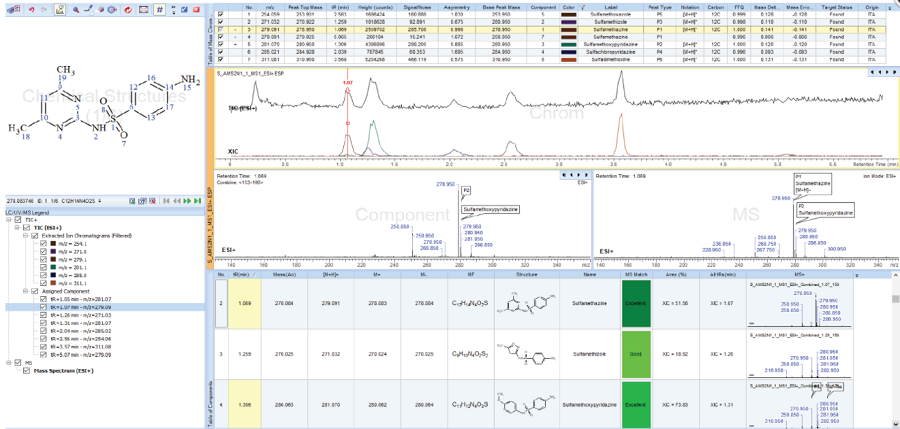
크로마토 그램을 추출하고 성분 목록을 스크리닝하여 알려진 화합물과 "알려진 미지" 화합물을 식별합니다.
MetaSense
Metabolite Identification that Just Makes Sense
시간을 절약하고 협업을 강화하면서 기존 대사 산물 식별 문제를 해결합니다.
- 대사 산물 예측 및 식별 자동화
- 생체 번환지도 및 보고서 자동 생성
- MetID 데이터를 추가하여 쉽게 대화 형의 검색 가능한 데이터베이스로 확장
- MetaSense 웹 포털을 통해 결과에 쉽게 접근하고 보고서 공유
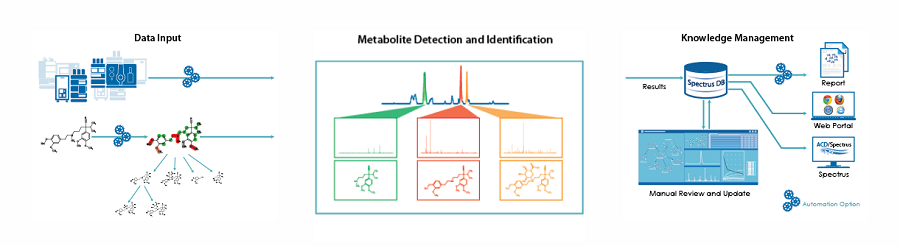
Method Selection Suite
Develop Better Chromatography Methods, in Less Time
물리화학적 성질 예측과 분리 방법 최적화 툴을 연계하여 방법 개발을 간소화하고 효과적으로 강력한 분리 방법을 생성합니다.
- 우세한 시작 조건을 정의하기 위해 성분들의 물리화학적 성질들을 예측합니다.
- Quality by Design (QbD) 원칙에 따라 분리 방법을 향상시키기 위해 다양한 실험 조건들을 시뮬레이션합니다.
(디자인 공간을 확장하는 다 변수 최적화)
- 사전 정의된 다양한 모델을 사용하여 분리 방법을 최적화합니다. (Proteins, HILIC 등)
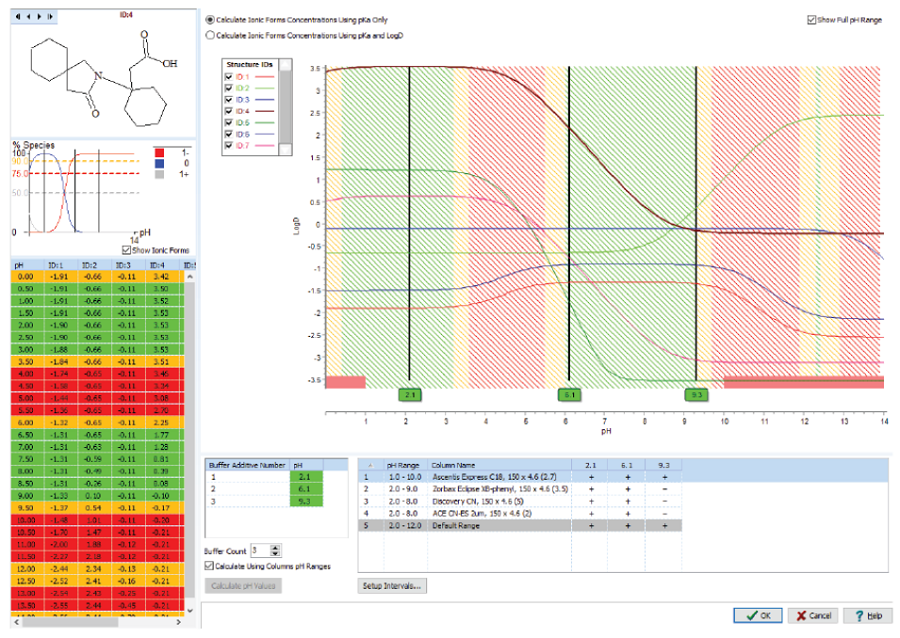
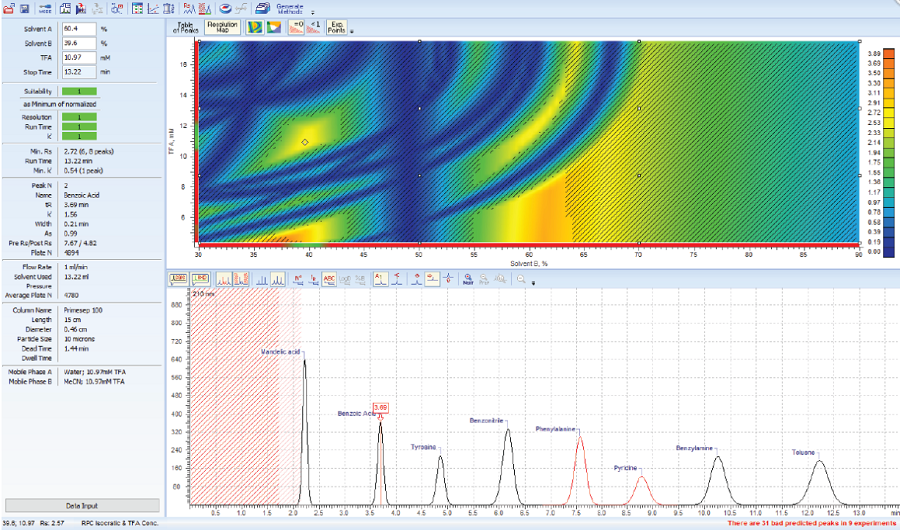
AutoChrom
Optimal Method Development Strategy
Quality by Design (QbD) 원칙에 따라 논리적 분석법을 개발하는 소프트웨어 패키지로 AutoChrom을 이용하여 최적의 LC/MS, LC/UV 분리 조건을 찾을 수 있습니다.
좋은 조건의 시작점을 선택하고 지능적인 최적화를 수행하여 짧은 시간에 더 나은 방법을 얻을 수 있습니다.
AutoChrom의 QbD 접근 방법은 다변수 최적화, 실험 공간 설계에 대한 체계적인 설계 방법을 제공하고 방법 개발 프로세스 전반에 걸친
견고한 모델링을 제공합니다.